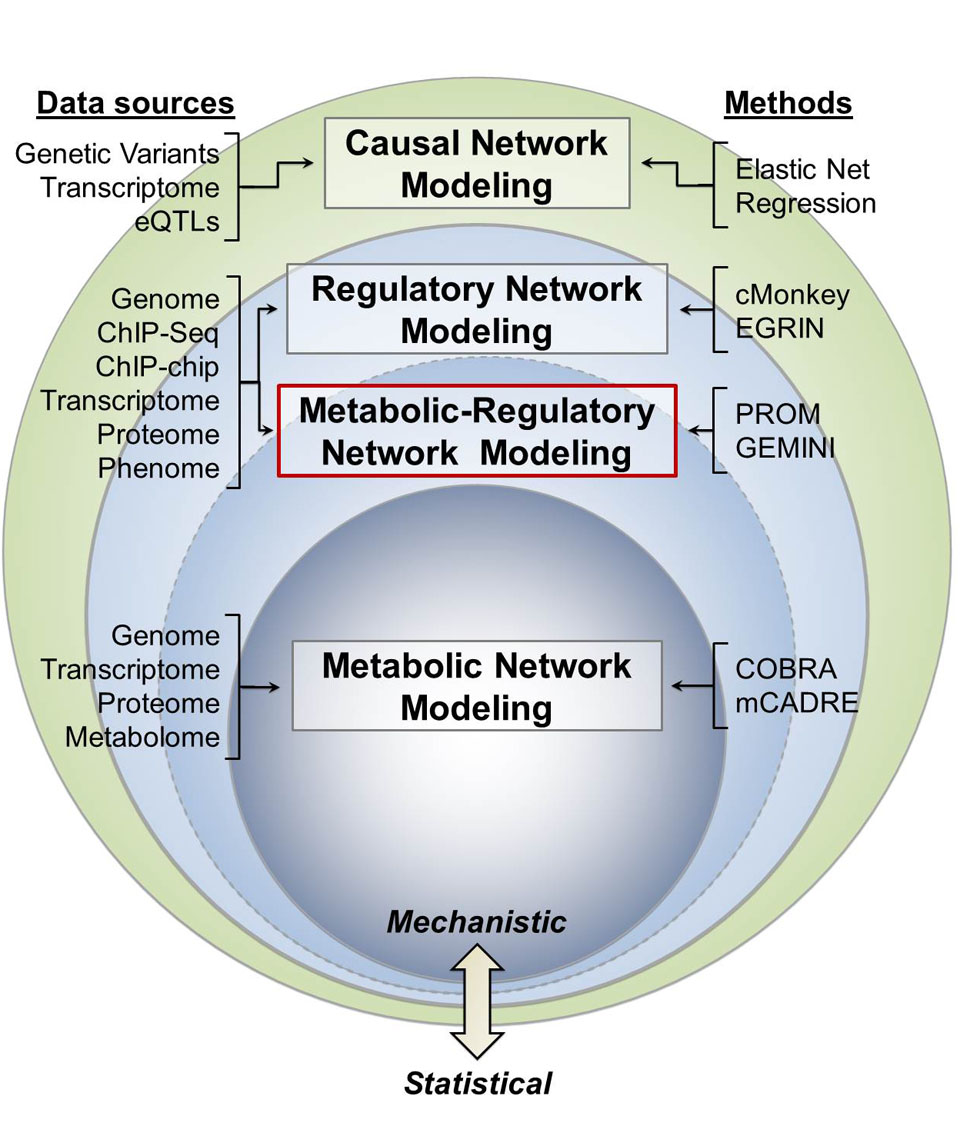
Integrated Network Modeling
High-throughput experimental technologies have enabled the identification and quantification of biological components at unprecedented scale, from genes and proteins to cells and tissues. Collectively, these technologies provide a parts list for biological systems. Computational systems biology employs an integrative approach to characterize biological systems, in which interactions among all components in a system are described mathematically to establish a computable model that cohesively integrates data.
For multiple reasons, biochemical networks of metabolism present an ideal core upon which to build integrated network models. For one, metabolic networks themselves integrate across multiple omics, including proteomics, transcriptomics, and metabolomics. While statistically inferred regulatory networks can portray relationships between environment, genotype, and expression state, metabolic networks provide direct, quantifiable connections between genotype and phenotype. As such, integrating these different scales of models is essential to more fully and accurately capture the scope of molecular processes in a biological system. As metabolic network models are grounded in detailed biochemistry, they are subject to physical and chemical laws, thus establishing valuable mechanistic context for statistical associations derived from top-down, data-driven approaches.
Predicting the response to genetic or environmental perturbations
Transcriptional regulation plays a key role in controlling metabolism and a forefront challenge in modeling organisms today is to build integrated models of regulation and metabolism. Predicting the effect of transcriptional perturbations on the metabolic network can lead to accurate predictions on how genetic mutations and perturbations are translated into flux responses at the metabolic level. The Probabilistic Regulation of Metabolism (PROM) algorithm that we developed represents the successful integration of a genome scale transcriptional regulatory network with a biochemically detailed metabolic network, bridging two important classes of systems biology models that have rarely been combined quantitatively.
The construction of an integrated metabolic-regulatory network using PROM requires the following: 1) the reconstructed genome scale metabolic network 2) regulatory network structure, consisting of transcription factors (TF) and their targets 3) gene expression data. We have used PROM to build genome-scale models for various model organisms and have shown that PROM can detect drug targets, identify gene knockout phenotypes with accuracies as high as 95% and predict microbial growth-rates of transcription factor knockout strains quantitatively with a correlation of 0.95.
Automated integration of regulatory and metabolic network models
GEMINI (Gene Expression and Metabolism Integrated for Network Inference) takes a draft regulatory network and then integrates it with the corresponding metabolic network using PROM (see above). It performs in silico knockouts of each transcription factor (TF) in the integrated model and compares the predictions with experimental observations. GEMINI identifies and removes interactions that do not lead to the measured growth phenotype, while retaining the phenotype-consistent interactions. This process is repeated iteratively for all TFs regulating metabolism in the network, yielding a set of phenotype-consistent interactions and a refined metabolic-regulatory model that is simultaneously consistent with observed gene knockout phenotypes, gene expression data, and the corresponding metabolic network state.


 hood-price.isbscience.org/research/integrated-network-modeling/
hood-price.isbscience.org/research/integrated-network-modeling/